2+2
1:100
3*3
sample(1:9)
X <- matrix(sample(1:9), nrow = 3, ncol = 3)
X
sum(X)
mean(X)
?sum
sum
nucleotides <- c("a", "c", "g", "t")
nucleotides
sample(nucleotides, size = 100, replace = TRUE)
table(sample(nucleotides, size = 100, replace = TRUE))
paste0(sample(nucleotides, size = 100, replace = TRUE), collapse = "")
replicate(n = 10, expr = paste0(sample(nucleotides, size = 100, replace = TRUE), collapse = ""))
df <- data.frame(id = paste0("seq", 1:10), seq = replicate(n = 10, expr = paste0(sample(nucleotides, size = 100, replace = TRUE), collapse = "")))
df
str(df)
ls()Lab 1: Course Intro & the Very Basics
Package(s)
Schedule
- 08.00 - 08.15: Arrival, fill in the pre-course anonymous questionaire and interest based group formation
- 08.15 - 08.45: Lecture: Course Introduction
- 08.45 - 09.00: Break
- 09.00 - 11.15: Exercises
- 11.15 - 11.30: Break
- 11.30 - 12.00: Lecture: Reproducibility in Modern Bio Data Science
Learning Materials
Please prepare the following materials:
- Read the full course description here: 22100 BSc / 22160 MSc
- If you have not already done so, please take 2 min. to answer the pre-course anonymous questionaire
- Read course site sections: Welcome to R for Bio Data Science, Prologue and lastly Getting Started, where it is important that you perform any small tasks mentioned
- Book: R4DS2e: Welcome
- Book: R4DS2e: Preface to the second edition
- Book: R4DS2e: Introduction
- Book: R4DS2e: Chapter 2 Workflow: basics
- Book: R4DS2e: Chapter 28 Quarto (Don’t do the exercises)
- Video: RStudio for the Total Beginner
- Paper: A Quick Guide to Organizing Computational Biology Projects
Unless explicitly stated, do not do the per-chapter exercises in the R4DS2e book
Learning Objectives
A student who has met the objectives of the session will be able to:
- Master the very basics of R
- Navigate the RStudio IDE
- Create, edit and run a basic Quarto document
- Explain why reproducible data analysis is important, as well as identify relevant challenges and explain replicability versus reproducibility
- Describe the components of a reproducible data analysis
Exercises
Today, we will focus on getting you started and up and running with the first elements of the course, namely the RStudio IDE (Integrated Developer Environment) and Quarto. If the relationship between R and RStudio is unclear, think of it this way: Consider a car, in that case, R would be the engine and RStudio would be the rest of the car. Neither is particularly useful, but together they form a functioning unit. Before you continue, make sure you in fact did watch the “RStudio for the Total Beginner” video (See the Learning Materials for today’s session).
First steps
Up and Running with the Course Cloud Server
In the menu on your left, you will find, amongst other things, some guides. Find the Guide for Cloud server and the RStudio IDE, click it and follow the guide.
The Console
Now, in the console, as you saw in the video, you can type commands like:
Take some time and play around with these commands and other things you can come up with. Use the ?function to get help on what that function does. Be sure to discuss what you observe in the console. Do not worry too much about the details for now; we are just getting started. But as you hopefully can see, R is very flexible and basically the message is: “If you can think it, you can build it in R”.
- Go to R4DS2e Chapter 2 Workflow: basics in R4DS2e and do the exercises
The Terminal
Notice how in the console pane, you also get a Terminal, click and enter:
ls
mkdir tmp
touch tmp/test.txt
ls tmp
rm tmp/test.txt
rmdir tmp
ls
echo $SHELLBasically, here you have access to a full terminal, which can prove immensely useful! Note, you may or may not be familiar with the concept of a terminal. Simply think of it as a way to interact with the computer using text command, rather than clicking on icons etc. Click back to the console.
The Source
The source is where you will write scripts. A script is a series of commands to be executed sequentially, i.e. first line 1, then line 2 and so on. Right now, you should have a open script called Untitled1. If not, you can create a new script by clicking white paper with a round green plus sign in the upper left corner.
Taking inspiration from the examples above, try to write a series of commands and include a print()-statement at the very end. Click File \(\rightarrow\) Save and save the file as e.g. my_first_script.R. Now, go to the console and type in the command source("my_first_script.R"). Congratulations! You have now written your very first reproducible R program!
The Whole Shebang
Enough playing around, let us embark on our modern Bio Data Science in R journey.
- In the
Filespane, clickNew Folderand create a folder calledprojects - In the upper right corner, click where it says
Project: (None)and then clickNew Project... - Click
New Directoryand thenNew Project - In the
Directory name:, enter e.g.r_for_bio_data_science - Click the
Browse...button and select your newly createdprojectsdirectory and then clickChoose - Click
Create Projectand wait a bit for it to get created
On Working in Projects
Projects allow you to create fully transferable bio data science projects, meaning that the root of the project will be where the .Rproj file is located. You can confirm this by entering getwd() in the console. This means that under no circumstances should you ever not work within a project, nor should you ever use absolute paths. Every single path you state in your project must be relative to the project root.
But why? Imagine you have created a project, where you have indeed used absolute paths. Now you want to share that project with a colleague. Said colleague gets your project and tests the reproducibility by running the project end-to-end. But it completely fails because you have hardcoded your paths to be absolute, meaning that all file and project resource locations point to locations on your laptop.
Projects are a must and allow you to create reproducible encapsulated bio data science projects. Note, the concept of reproducibility is absolutely central to this course and must be considered in all aspects of the life cycle of a project!
If projects and paths seem unfamiliar to you, you may want to look in the menu on the left and find the Primers and then find and click Paths & Projects, which will guide you through the details.
Quarto
While .R-scripts are a perfectly valid way to write scripts, there is another Skywalker:
- In the upper left corner, again, click the white paper with the round green plus, but this time select
Quarto Document - Enter a
Title:, e.g. “Lab 1 Exercises” and enter your name below in the boxAuthor: - Click
Create - Important: Save your Quarto document! Click
File\(\rightarrow\)Saveand name it e.g.lab_01_exercises.qmd - Minimise the
Environmentpane
You should now see something like this:
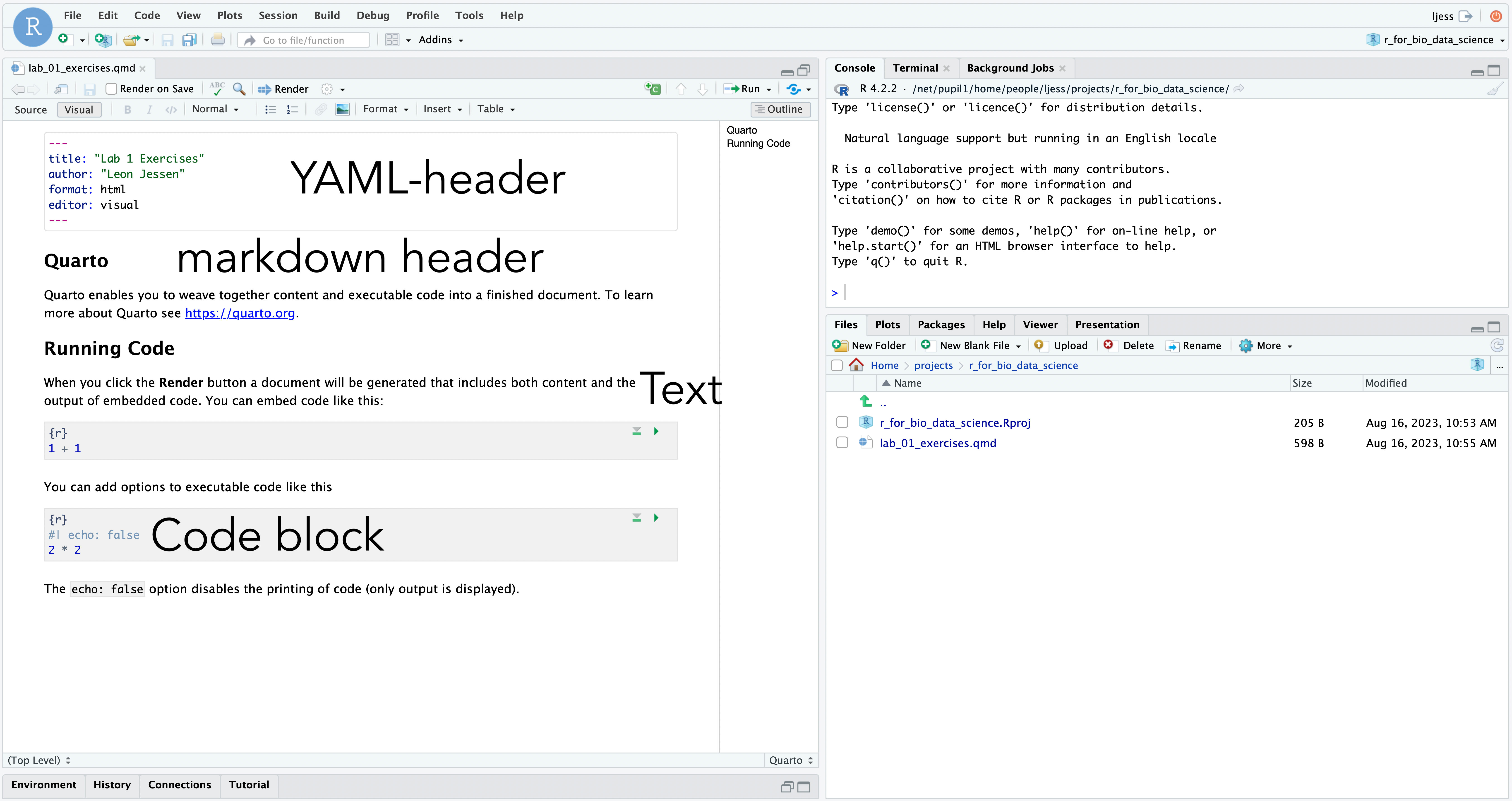
Try clicking the Render button just above the quarto-document. This will create the HTML5 output file. Note! You may get a Connection Refused message. If so, don’t worry, just close the page to return to the cloud server and find the generated .html-file, left-click and select View in Web Browser.
You may also get a bit of a weird message, which could look something like this /file_show?path=%2Fnet%2Fpupil1%2Fhome%2Fpeople%2Fztizh%2Ftry.html not found. In that case, click the generated .html-file and choose Open in Editor and then at the top of document, find the button saying Preview and click it. Note, this is a work-around.
If you have previously worked with Rmarkdown, then many features of Quarto will be familiar. Think of Quarto as a complete rethinking of Rmarkdown, i.e. based on all the experience gained, what would the optimal way of constructing an open-source scientific and technical publishing system?
If you have previously encountered Jupyter notebooks, Quarto is similar. The basic idea is to have one document covering the entire project cycle.
Proceed to R4DS2e Chapter 28 Quarto and do the exercises.
Artificial Intelligence in modern Bio Data Science
This section is new as of 2025, please read carefully and follow along…
Honestly, if AI is going to solve all coding and analysis tasks moving forth, then why even bother??? 🤷
This technology has made its entrance and is here to stay, but in order to really increase your learning yield, it is very important to understand how to use this technology.
You can compare these technologies with being in the same group as the absolute smartest students in class, in fact the smartest student who has ever taken this class.
It is really important to distinguish the following two scenarios:
- Getting the smart student to solve your programming/analysis task
- Getting feedback from the smart student on how you solved your programming/analysis task
In scenario 1, you are robbed of your thought processes on how to approach the task, divide into to manageable sub tasks and then come up with structured solutions, which combined, will solve the overall task. This process is referred to as “Computational Thinking”.
In scenario 2, you go through the above outlined computational thinking process and you train and develop your competencies
In this course, the importance of being aware of actively building and training your computational thinking competencies, while also building your technical toolbox cannot be underestimated!!!
Can an AI solve the majority of tasks you will work with in this course? YES!!! BUT, it is up to you as a student to actively engage the course taking responsibility for building competencies as outlined above.
Basically, what will you learn, if you let the smartest student do your homework? 🤷
Here is the long term catch, if you do not possess competencies beyond what an AI can do, then why would a future employer hire you instead of just getting an AI to solve the tasks???
It is not just about passing this course, it is about understanding that in this course, you will be building the foundation for your future career.
Getting started
For this exercise, you will need access to some
There are many others and more keep coming, the development in this field is extremely rapid!
In the following, please feel free to use whichever of the above you prefer or perhaps try one, which is new to you.
Let us get acquainted
I am assuming that most students by now have encountered this technology. Nevertheless, let us get this exercise started:
- T1: Start a new chat and enter the question: “Explain in simple terms what you are”
- T2: Now enter the question: “Explain in simple terms how you work”
And then in your group, compare the output from the AI and answer:
Q1: Were the answers the same, when using different AIs?
Q2: Were the answers the same, when using the same AI?
Q3: Discuss in your group why/why not?
T3: Use your favourite AI and try to get input on your “why/why not”-discussion
Q4: Discuss in your group if the AI added additional reflections on why/why not and what the consequence of this is in context with solving tasks?
Q5: Discuss in your group: Will the same person always give the same answer? How about different persons?
Q6: Discuss in your group if these AIs basically are human? Discuss how they are created, how they generate text and if this limits their capabilities compared to humans. If you do not know how they are created of how the generate text, ask them
Using AI as an assistant, a use case
In this use case, we will work with the following paper:
You are working as a researcher, or a consultant or a bioinformatician or a bio data scientist and your boss have given you this paper and asked you to look into it. You decide to use AI:
- T4: Download the paper, then upload it to your favourite AI and ask it: “Explain in simple terms what this paper is a about and create a simple brief summary” and read the summary, perhaps if something is unclear, ask the AI to elaborate
You take interest in the pseudo-sequence distance metric and again you decide to use AI:
- T5: Ask your favourite AI: “Please in simple terms explain the pseudo-sequence distance metric used in the paper”, again if something is unclear, simply ask for elaboration
Likely your AI will outline something concerning similarity or distance between these protein molecules. Let us dig a bit further in:
- T6: Ask your favourite AI: “Please in simple terms explain exactly how the authors calculate this pseudo-sequence distance metric between two molecules”
The AI may or may not point you in the right direction. Being a bit unsure of this, you dig into the paper yourself and you find equation 2:
- T7: Ask your favourite AI: “As I read the paper, the authors define the pseudo-sequence distance metric using equation 2. Please state the equation and in simple terms explain the terms”
Again, the AI may or may not point you in the right direction.
- T8: Check that the equation the AI returns looks like equation 2, if not (which may or may not happen!):
- T8a: Take a moment to ponder: You were quite specific in your instructions and the AI returned a wrong equation with FULL confidence! (This is called AI hallucination and it’s something to be VERY aware of!)
- T8b: Then state: “Looking at the equation you have stated, I see it does not seem to match equation 2 in the paper. Please check and correct”
Once you are confident that the AI has the right equation you now want to impress your boss and decide to create an R-function:
- T9: Check that the equation the AI returns looks exactly like equation 2 and then state: “Please propose an R-function, which given two pseudo-sequences computes and returns the pseudo-sequence distance metric. Include an example of running the function and getting a result”
Note, in the proposed code, you can simply in the upper right corner click copy.
- T10: Copy the code and paste it into the console in your RStudio Cloud server session and then run it, by hitting return. Run the function with these two examples
CASSIRSSYEQYFandCASSLGQGNTLYFand compare with your group, did all of your functions run? Did you all get the same value?
Pssst… Don’t know how to do this and need a hint? Try to click here…
Well, in this exercise, ask your faourite AI: “How do I run these two examplespaste examples through this function paste function
- T11: Now, return to your favourite AI and enter: “Please propose an R-function for randomly generating a pseudo sequence. Then use that to compute the pseudo-sequence distance metric 10 times”
Again, run this code and check the 10 computed distances. The exact meaning of these 10 examples is not clear to you, so you decide to again turn to your AI:
- T12: Enter: “The exact meaning of these values are not clear to me. Here are my computed values:” and then copy paste the computed values from your R-console
The AI will now come with some longer explanation, which may or may not be correct.
Ok, so now you want to collect everything, so you enter: “I now want a complete script, please create a script, with a function implementing the pseudo sequence distance calculation, a function, which can randomly generate amino acid sequences and then finally, generate 2 times 10 random amino acid sequences and compute all pseudo sequence distances”
And then let’s vibe-code: Simply copy/paste the script and run it in the console, if it does not run, simply copy/paste the error into your favourite AI and keep doing that until you code runs.
- T13: Once your code runs, try to see if you can share your examples with your group and compare if you get the same distances
Finally:
Q7: Assuming your code runs, discuss if you understand the details of the code and whether you would be comfortable sharing the code into an environment, where it is to support a multi-million-dollar drug development pipeline?
Q8: Knowing that these AI assistants can hallucinate, i.e. give you information, which is completely wrong equally confident as information, which is correct, how do you know, if your function does what it is supposed to?
That’s it for today and remember moving in this course: DON’T use AI to generate the solution to an exercise, instead think your way through the exercise, generate YOUR solution and then DO get feedback from AI on your solution!
Put in the time and the effort in this course and you will learn how to create the good solution and often even better than an AI and that’s your angle right there!